Performance Comparison of Polars vs Pandas
Is Polars really faster? How fast is it? How do the different syntaxes perform?
If you haven't read it already, do check out my article Introducing Polars, which goes over what is Polars, who it is for and the differences from Pandas in detail. This article only briefly touches upon them and focuses on the performance comparison,
🐻❄️ Introduction to Polars
Polars is an open-source data manipulation library that offers faster processing speeds and efficient memory usage compared to Pandas. It is built in Rust, a programming language known for its speed and safety, and offers a DataFrame API that is similar to Pandas.
When working with large datasets, choosing the right data manipulation library can significantly affect performance. In this blog, we will compare the performance of two popular data manipulation libraries: Pandas and Polars, using benchmarking.
🐼 Differences between Pandas and Polars
Pandas is a widely used data manipulation library that offers a comprehensive set of tools for data analysis. However, its performance is limited by its reliance on the Python programming language, which is known to be slower than languages like Rust. In contrast, Polars is built in Rust, which enables it to deliver significantly faster performance.
One of the key differences between Pandas and Polars is their memory usage. Pandas stores data in memory as a NumPy array, which can be memory-intensive for large datasets. On the other hand, Polars uses memory mapping, which enables it to read and write data to disk without loading it into memory. This results in more efficient memory usage, especially for large datasets.
Another significant difference between Pandas and Polars is their processing speed. Polars is designed to leverage the power of modern CPUs, making use of multi-threading and SIMD (Single Instruction Multiple Data) instructions. This enables it to perform operations like aggregations, joins, and filters much faster than Pandas, especially on large datasets.
🏄 Set Up
The setup is simple, we need to install pyarrow and polars. Without pyarrow, polars complains about it not being present. The experiments are run on an M1 Mac with 16GB ram and 256 GB SSD.
We will create a virtualenv for the experiment and run
pip install polars pandas pyarrow
To begin with, we create a data frame with 1 million rows and 10 columns, using NumPy's random function. We will use this data frame for our benchmarking.
# Create a datafram with 1 million rows and 10 columns
df = pd.DataFrame(np.random.rand(10000000, 10), columns=['col_'+str(i) for i in range(10)])
pl_df = pl.DataFrame(df)
🔔 Brief aside on %timeit
The %timeit magic command is used in Jupyter notebooks to measure the time taken by a piece of code to execute. The command runs the code multiple times to get an average execution time, which helps in eliminating any variations in the time taken by the code to execute due to system performance or other factors.
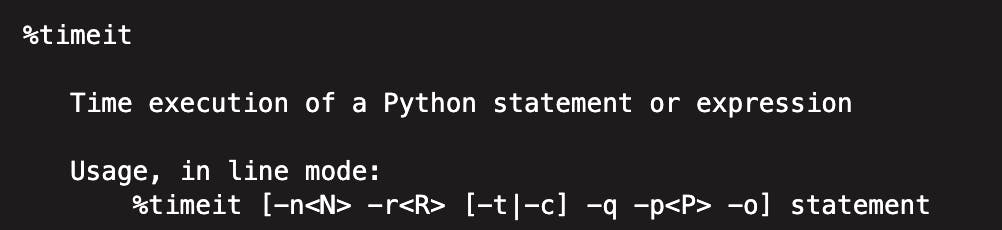
In the code snippets provided below, %timeit is used to compare the performance of Pandas and Polars libraries for various operations. The -r7 parameter, for example, indicates that the command should run the code 7 times, and -n 1000 specifies that the command should run the code 1000 times for each run. This ensures that the results obtained are statistically significant and reliable.
By using %timeit in this way, we can compare the performance of Pandas and Polars for various operations and determine which library is faster and more efficient for each command.
Now let's get into the operations we're going to benchmark.
📊 Benchmarking
📈 Selecting Columns
We start our benchmarking by selecting a column. We select columns 'col_0' and 'col_1' from our data frame using Pandas and Polars.
# Selecting columns
#Pandas
%timeit -r7 -n 1000 df[['col_0','col_1']]
#Polars
%timeit -r7 -n 1000 pl_df[['col_0','col_1']]
%timeit -r7 -n 1000 pl_df.select(pl.col(['col_0','col_1']))
The above code measures the execution time for each of the three operations (Pandas, Polars - using square brackets, and Polars - using select method) for 7 rounds with 1000 executions each.
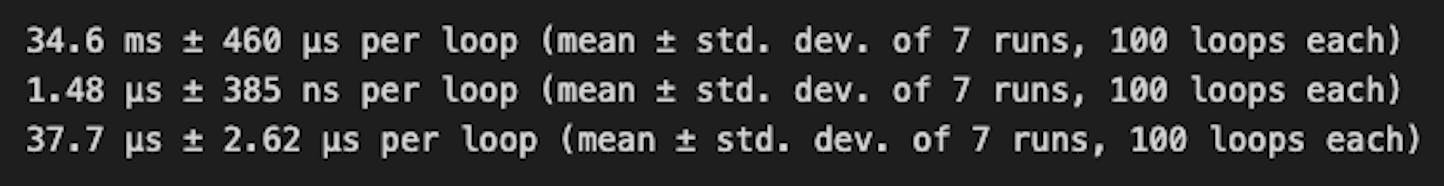
Pandas took 34.6ms, whereas Polars took 1.48μs with indexing and 37.7μs for the select method.
The results show that Polars outperforms Pandas significantly for this operation, and highlight that square bracket indexing performed ~25 times faster than the select API for filter, which is generally the recommended mode. This exception is also called out in the polars docs, selecting with indexing section.
Polars select API (37.7μs) is ~1000 times faster than Pandas (34.6ms).
Polars square bracket indexing (1.48μs) is about 23,000 times faster than Pandas (34.6ms).
📈 Filtering Rows
Next, we filter rows based on a condition. We select rows where 'col_0' is greater than 0.5 using Pandas and Polars.
#Filtering rows
#Pandas
%timeit -r7 -n 1000 df.query ('col_0>0.5')
#Polars
%timeit -r7 -n 1000 pl_df.filter(pl.col('col_0')>0.5)

The results show that Polars is faster than Pandas for this operation as well, but the magnitude is far lesser.
Pandas (108ms) takes about 2.8 times as long as Polars (38.3ms) in filtering rows.
📉 Groupby
In the next operation, we group by 'col_0' and calculate the mean of 'col_1'. We use both Pandas and Polars for this operation.
#Grouping by col_0 and calculating the mean of col_1
%timeit -r7 -n 1000 df.groupby('col_0')['col_1'].mean()
%timeit -r7 -n 1000 df.groupby('col_0')['col_1'].agg('mean')
#polars
%timeit -r7 -n 1000 pl_df.groupby('col_0').agg([pl.col('col_1').mean()]) #select method
%timeit -r7 -n 1000 pl_df.groupby('col_0').agg(pl.mean('col_1'))#short
The results show that Polars is slower than Pandas in doing groupby. Polars offers two syntaxes for this operation, and both are faster than Pandas.
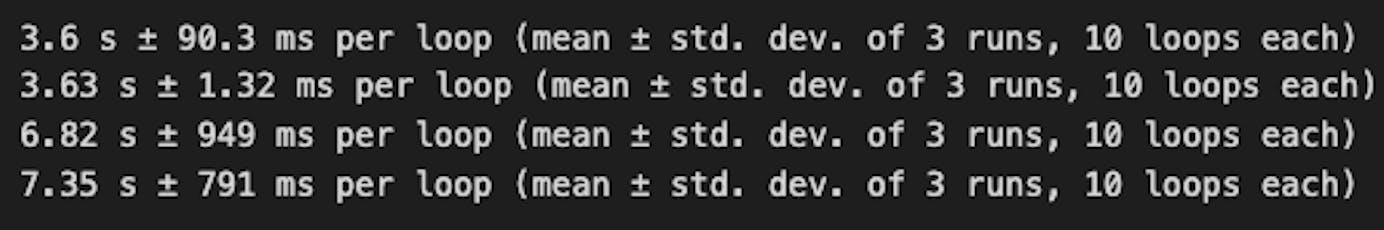
In Pandas, .mean() was slightly slower than .agg('mean')
In Polars, using the pl.col function to extract the column and then calling .mean() was slightly faster than calling pl.mean directly within square indexing on the column.
Overall, Pandas (3.62s) was almost twice as fast as Polars(7s), irrespective of the syntax used.
📈 Conversion between wide and long formats
Lastly, we compare the performance of Pandas and Polars for conversion between wide and long formats using the melt function.
#Conversion between wide and long formats
#Pandas
%timeit -r7 -n 1000 pd.melt(df, id_vars=['col_0'], value_vars=['col_1', 'col_2', 'col_3', 'col_4', 'col_5', 'col_6', 'col_7', 'col_8', 'col_9'])
#Polars
%timeit -r7 -n 1000 pl_df.melt(id_vars=['col_0'], value_vars=['col_1', 'col_2', 'col_3', 'col_4', 'col_5', 'col_6', 'col_7', 'col_8', 'col_9'])

Thus Polars (696ms) is about than 5.5 times faster than Pandas (3.88s) for melt.
🎉 Wrap Up
In conclusion, Polars offers a compelling alternative to Pandas for data manipulation tasks, particularly when dealing with large datasets that require efficient memory usage and high processing speeds. While Pandas remains a popular and powerful library, Polars' performance advantages make it a valuable addition to any data scientist or data engineer's toolkit.
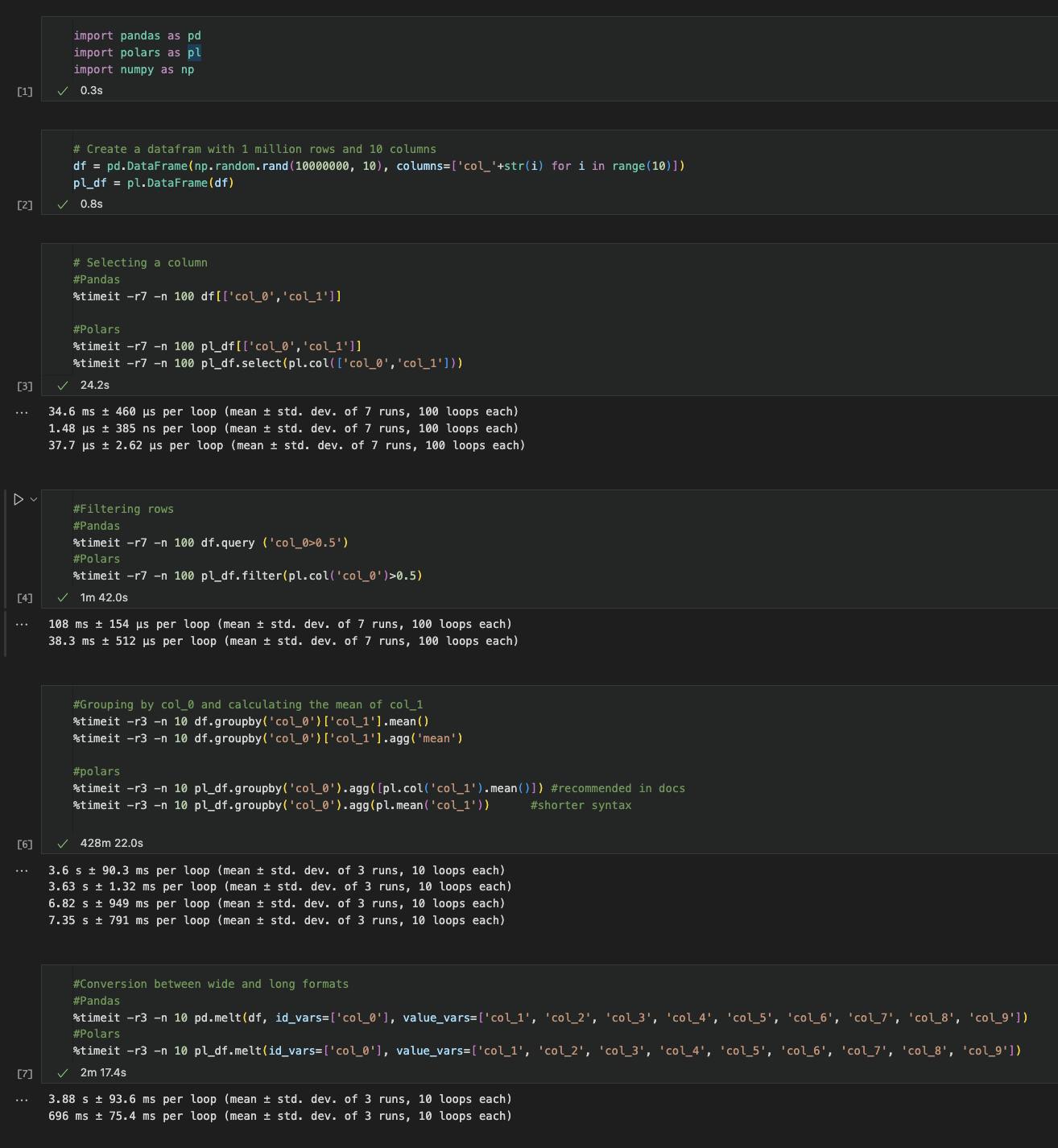
Here's an image with all the code and runtimes together.
I highly recommend reading the official Polars User Guide, if you're loving this series to learn more about the syntax, and query optimization. I'd love to hear on social media, which Polars feature or feature set you want me to cover in the next article.
If you liked this article, please show some love and do share it with your network who would find it valuable. See you in the next article.

